I am almost done with the first big update for our DoOR database. I received and integrated tons of data from colleagues all over the world and I updated, wrote and rewrote endless lines of code. Now I finally have the time to polish here and there, e.g. update our plotting functions. Right now I am working on the new antennal lobe (AL) activity maps. We use these maps in DoOR to visualize the ensemble responses each individual odorant elicits across all Drosophila sensory neurons. For example the activity pattern for Isopentyl acetate looked like this in DoOR 1.0:
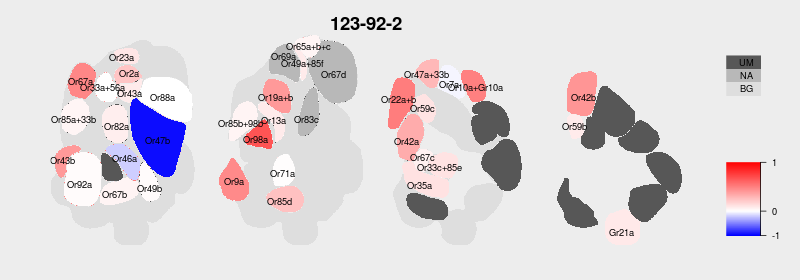
On the one hand I had to map the deorphanized glomeruli (dark grey, UM = unmapped). But there were also some glomeruli missing on this AL map. I asked Veit Grabe who recently published a great in-vivo 3D atlas [1] of the Drosophila AL if he had a better AL representation for me. Not only did he have one, he even created a beautiful 2D map that perfectly fitte our need of displaying all glomeruli in only a few sections.
Next I had to find a way to integrate this new map into DoOR. Technically the old AL map was a pixel graphic, existing as a matrix in R. Each glomerulus had a specific grey value, for mapping odorant responses we exchanged the grey values with the corresponding color-coded activity values. I always wanted to upgrade this pixel-map to a vector based map, for visual reasons and as mapping responses should be much more straight forward. But how to get SVG paths into R?
Importing SVG maps into R as polygon
There seems to be no easy way to get a SVG into R. I found one thread suggesting to parse the SVG using the XML package. But this did not produce useful results, at least in my hands. I was able to read in the points but something went terribly wrong, making the plots look like stars rather than glomeruli.
I ended up with a rather complicated method:
(My main sources were this and this. Unfortunately I can’t find the post anymore that explained how to convert the SVG to SHP via DXF using Inkscape and QGIS):
- First I had to export the SVG from Inkscape to some AutoCAD .dxf file.
- Next, the .dxf file could be imported into QGIS which is normally used to work with geographical data, but what is the AL other than a map?
- From QGIS the map could be saved as a shape file which seems to be a standard for geographical data. For some reason glomeruli were not recognized as entities when importing the DXF files. They were split into several segments (“features”) which I had to select and merge individually (you have to import dgf, then save as shape file by right clicking the imported layer and choose “save as”, then you have to re-import the .shp file as otherwise you cannot edit it. Next you have to toggle the edit mode, select features and hit “Merge Selected Features”… This procedure took me hours…).
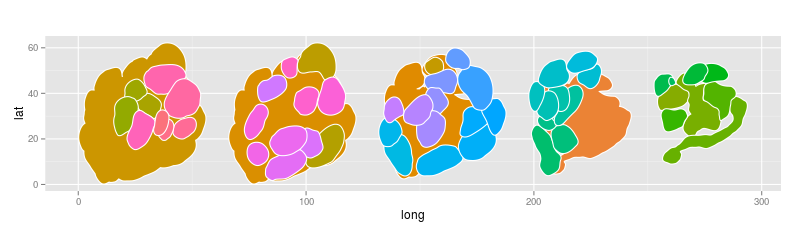
- In R I installed the rgl package, imported the shape file, removed empty groups and successfully plotted the AL with each glomerulus having assigned its own group number:
require(maptools)
require(rgdal)
require(ggplot2)
AL <- readOGR(dsn = "../ALimage/DoOR 2.0/", layer = "AL2polygon_finished")
AL <- fortify(AL, region="GEO_ID")
AL <- AL[- which(AL$group %in% levels(AL$group)[64:119]),] # remove empty groups
AL$group <- factor(AL$group, levels = rev(levels(factor(AL$group)))) #reverse order of levels
head(AL)
ggplot(data = AL) +
geom_polygon(aes(x = long, y = lat, fill=group), color = "white", size = .5) +
coord_fixed() +
theme(legend.position = "none")Much nicer right? Thank you Veit!
Whats left now is to adjust the z-arrangement of glomeruli-polygons, mapping the glomeruli and updating the functions for plotting.
References
-
Grabe, V., Strutz, A., Baschwitz, A., Hansson, B.S., Sachse, S., 2015. Digital in vivo 3D atlas of the antennal lobe of Drosophila melanogaster. Journal of Comparative Neurology 523, 530–544. doi:10.1002/cne.23697 ↩
